Struggling with sluggish searches? By leveraging PostgreSQL’s native full-text search, we cut query times from seconds to milliseconds, giving clinicians instant access to critical patient data.
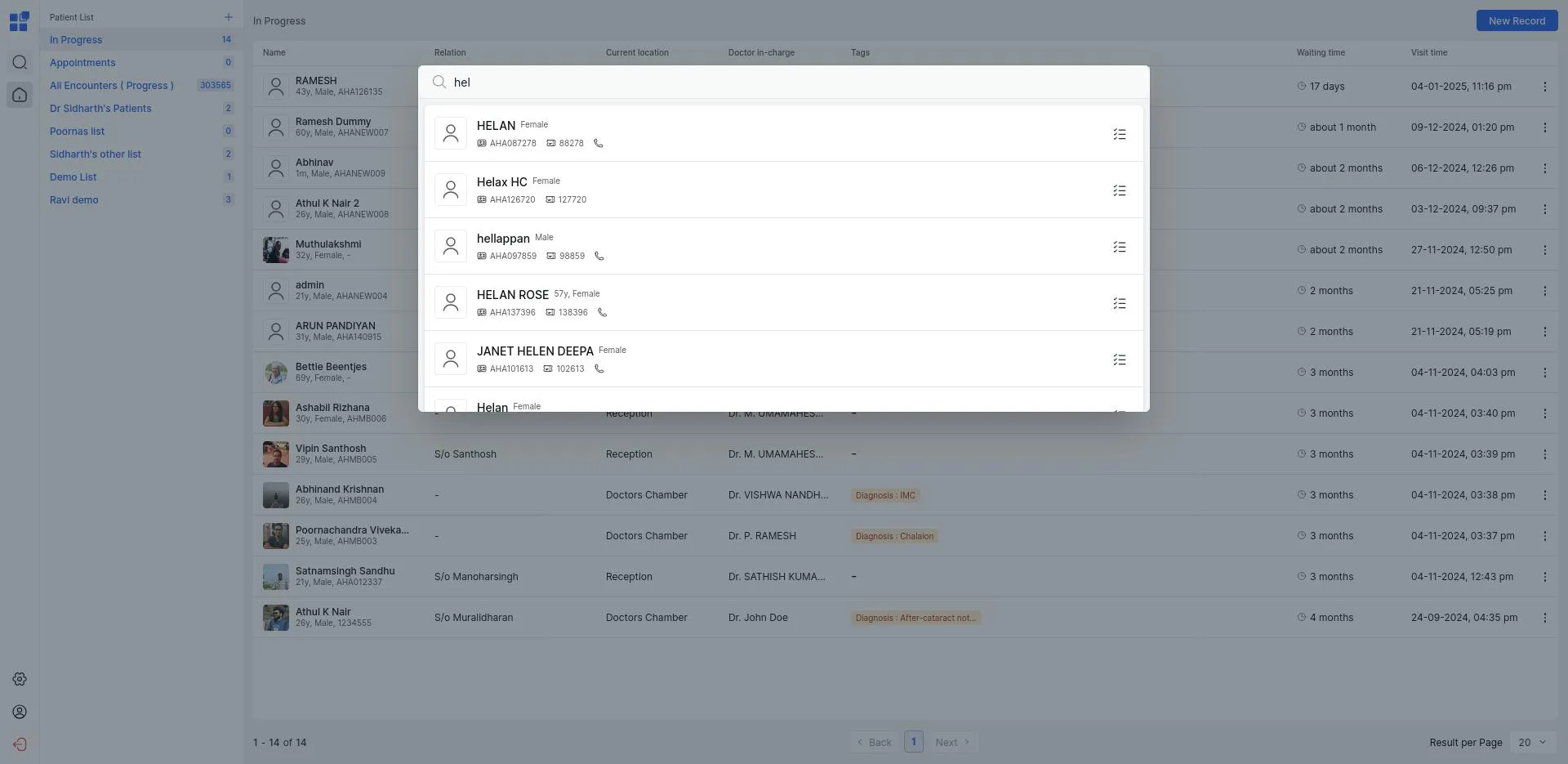 Searching for patients is an essential feature on our platform that gets used very very often during the course of the day on our platform. This search was implemented ontop of FHIR encounter and patient resources. By talking to our users we realised that there were three things they wanted to use by:
Searching for patients is an essential feature on our platform that gets used very very often during the course of the day on our platform. This search was implemented ontop of FHIR encounter and patient resources. By talking to our users we realised that there were three things they wanted to use by:
- Name
- Phone number
- Hospital UHID
When initially analyzing how we could deliver optimal search experience for our users we evaluated search engine options such as Elastisearch, Typesense and Millisearch. Then we reviewed Postgres’ text search and a few articles such as CrunchyData’s blog on Postgres that dived into it and realised that it could meet our expectations without adding another dependency to our stack.
Patient name search
We then wanted to evaluate performance of using Postgres native feature called tsvector and tsquery, its well documented on the documentation site: https://www.postgresql.org/docs/current/datatype-textsearch.html
Here is what we observed for our usecase:
name: [ { "text": "Athul" } ]This means that our queries resemble
EXPLAIN ANALYZE SELECT *
FROM fhir.patient
WHERE EXISTS (
SELECT 1
WHERE fhir.patient.name -> 0 ->> 'text' ILIKE '%Athu%'
);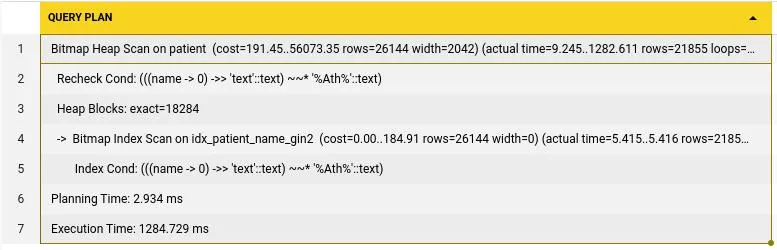 This query plan shows us that this query took 1284ms to execute and 2.9ms to plan, this means that planning took 68% of the whole time even with Trigram indexes.
This query plan shows us that this query took 1284ms to execute and 2.9ms to plan, this means that planning took 68% of the whole time even with Trigram indexes.
We then created a ts_vector column for the same column
ALTER TABLE fhir.patient
ADD COLUMN tsv_text tsvector GENERATED ALWAYS AS (
to_tsvector('english', COALESCE(name->0->>'text', ''))
) STORED;
CREATE INDEX idx_fhir_patient_tsv_text ON fhir.patient USING GIN (tsv_text);We then ran the analyze command to understand if this sped things up
EXPLAIN ANALYZE SELECT *
FROM fhir.patient
WHERE tsv_text @@ to_tsquery('english', 'Ath:*')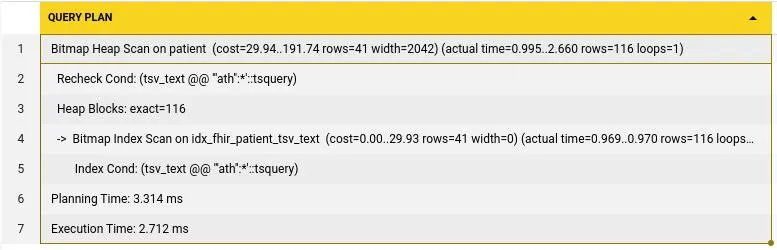 Execution time was reduced to just 2.7 ms from 1284ms and planning to 0.25ms. This means we saw a 475X reduction in execution time but with average 10X reduction compared to previous search times, all within postgres.
Execution time was reduced to just 2.7 ms from 1284ms and planning to 0.25ms. This means we saw a 475X reduction in execution time but with average 10X reduction compared to previous search times, all within postgres.
Identifier search
Each patient gets tagged with a unique identifier that we also need to search by, here is how store the identifier inside postgres jsonb[] column
[{"value": "119050", "system": "https://medblocks.com/fhir/new_id},
{"value": "AHA118050","system": "https://medblocks.com/fhir/uid"},
{"value": "33438-111019","system": "https://medblocks.com/fhir/uid_old" }]When we wanted to search by all the IDs we would unnest and then search each of them like so,
EXPLAIN ANALYZE SELECT *
FROM fhir.patient
WHERE EXISTS (
SELECT 1
FROM UNNEST(fhir.patient.identifier) AS elem
WHERE elem ->> 'value' LIKE ANY (ARRAY['%AHMB004%'])
)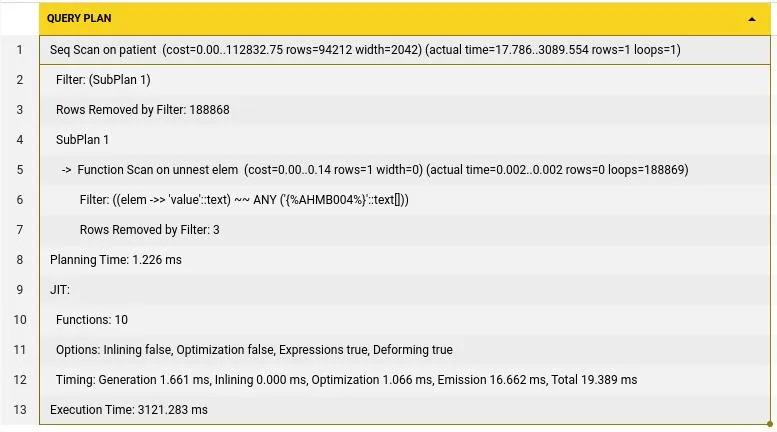 This search takes 3121ms to run, which is quite high. Now lets also include this into our ts vector column to see what the speed up it could result in. Lets write a trigger function since postgres GENERATED ALWAYS does not work with complex functions to update the field when identifiers are added.
This search takes 3121ms to run, which is quite high. Now lets also include this into our ts vector column to see what the speed up it could result in. Lets write a trigger function since postgres GENERATED ALWAYS does not work with complex functions to update the field when identifiers are added.
ALTER TABLE fhir.patient ADD COLUMN tsv_text tsvector;
CREATE INDEX idx_fhir_patient_tsv_text ON fhir.patient USING GIN (tsv_text);
CREATE OR REPLACE FUNCTION update_tsv_text()
RETURNS TRIGGER AS $$
BEGIN
NEW.tsv_text := to_tsvector(
'english',
COALESCE(NEW.name->0->>'text', '') || ' ' ||
COALESCE((
SELECT string_agg(elem->>'value', ' ')
FROM unnest(NEW.identifier) elem
), '')
);
RETURN NEW;
END;
$$ LANGUAGE plpgsql;
CREATE TRIGGER set_tsv_text BEFORE INSERT OR UPDATE ON fhir.patient FOR EACH ROW EXECUTE FUNCTION update_tsv_text();Lets implement the above and see what the cost implications are for searching both identifier and name Now the identifier search has a similar cost like the name search. Even a combination search of both name and identifier shows a lower execution time than earlier individual query.
EXPLAIN ANALYZE SELECT *
FROM fhir.patient
WHERE tsv_text @@ to_tsquery('english', 'Abhi:*')
AND tsv_text @@ to_tsquery('english', 'AHMB004:*')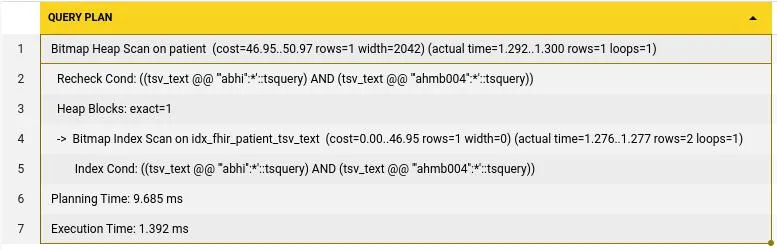 The cost of execution here is down to only 1.392ms from the initial 3121ms each search used to take. This is a huge gain with just built in postgres features.
The cost of execution here is down to only 1.392ms from the initial 3121ms each search used to take. This is a huge gain with just built in postgres features.
Real world implications
When the above changes were made in our product and deployed, we noticed a huge improvement in the search loading time which was anticipated. However the other effect that we saw an improvement in was CPU utilization.
After the implementation of these queries we found that the average cpu utilisation came down from 23.23 ms (23rd Dec 2024 to 3rd Jan 2025) to 6.13 ms (4th Jan 2025 to 18th Jan 2025).
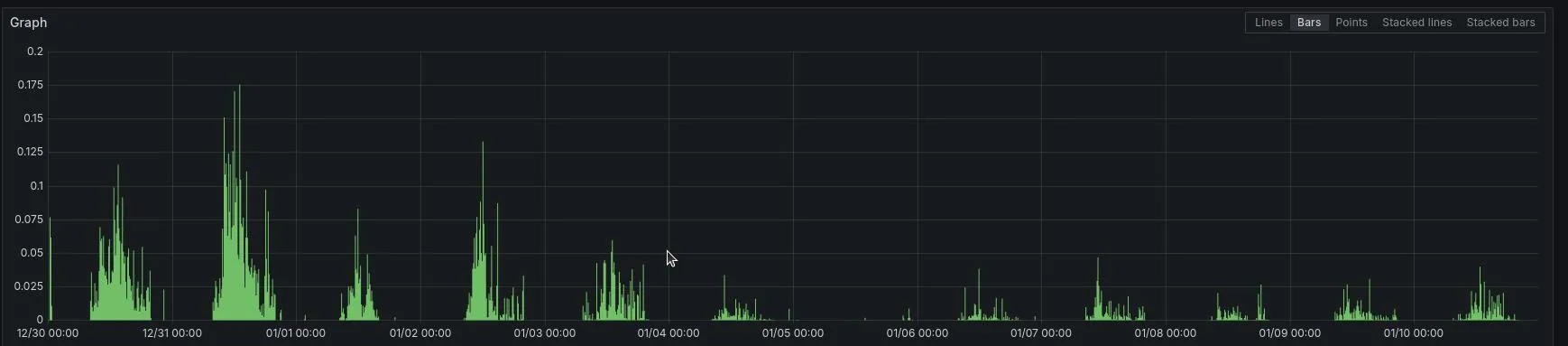 The above graph shows the CPU throttling for the postgres instance that contained our patient data. You can clearly observe a notable decrease in the peaks post 4th Jan when this was implemented.
The above graph shows the CPU throttling for the postgres instance that contained our patient data. You can clearly observe a notable decrease in the peaks post 4th Jan when this was implemented.
(Note: The peaks observed are actually the database backup job that runs at 2AM)
