Monday morning tumor board. A 58‑year‑old with a 7 cm mass, ER+/HER2−, nodes positive. The question in the room is what to do next, and how to do it safely. The attending opens up the EHR to reconstruct the story: he’s furiously opening up multiple windows to see imaging, core biopsy, pathology markers, staging, prior meds.
Someone pulls up the latest NCCN guidelines. It’s a PDF document on their email. After scrolling through the document, the flowchart for the current case looks something like this:
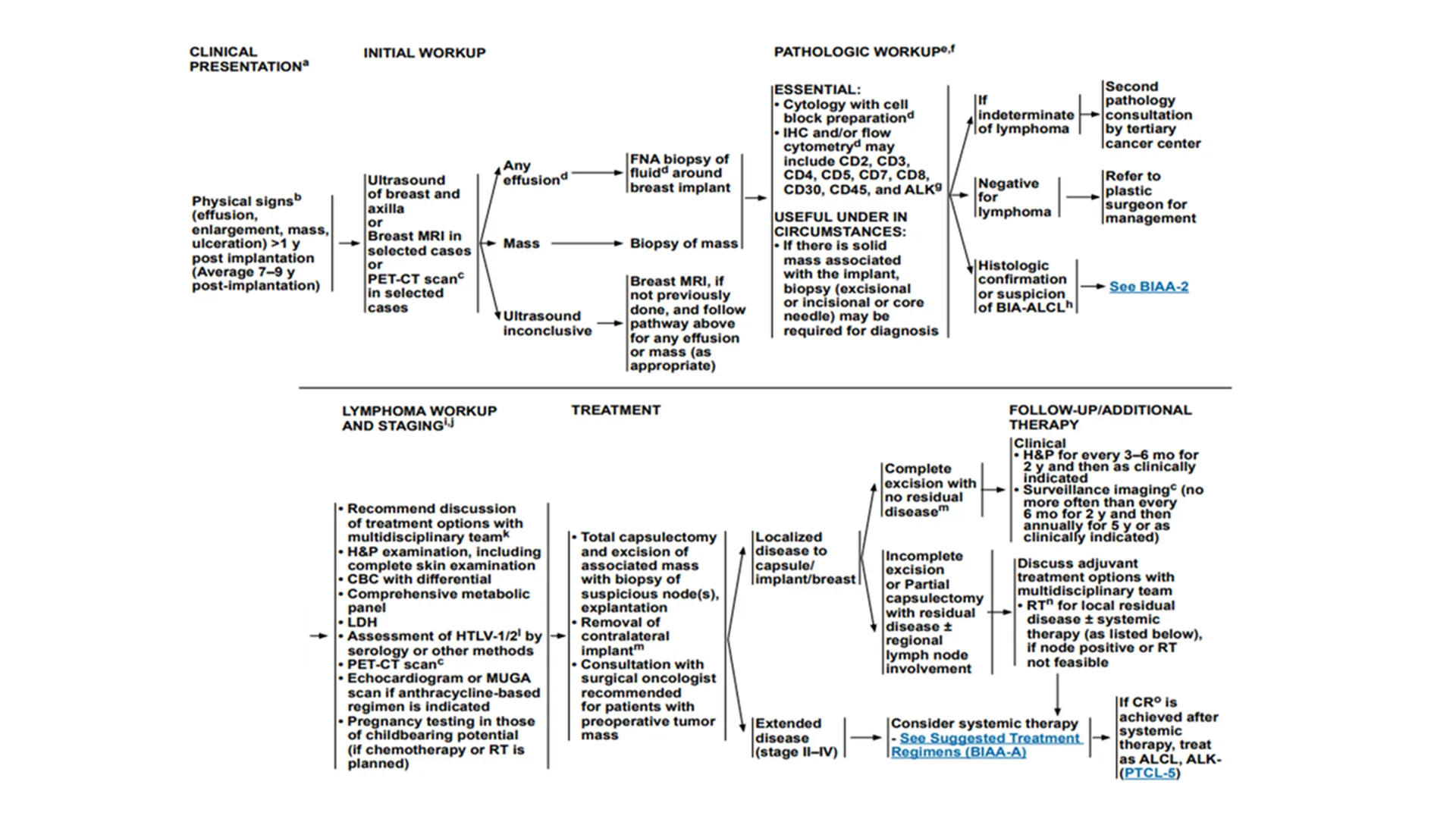
The team must translate boxes and arrows into orders and rationale. Tumor size is buried in radiology, immuno‑markers in scanned PDFs, staging in a note. Different departments label the same concept differently sometimes.
This is the average week for the tumor board: reconcile inconsistent data, map it to the guideline, then defend the regimen at tumor board. It’s slow, error‑prone, and the reasoning becomes invisible once the order is placed.
In 2023, GE HealthCare approached Medblocks to help them build an explainable Clinical Decision Support for the OncoCare product.

Introduction
GE HealthCare is a global leader in medical technology, diagnostics, and digital solutions. It became an independent company from GE after its spin‑off. (So technically, we worked with both GE and GE HealthCare during this engagement).
GE HealthCare is among the top three diagnostic imaging vendors, with about 18% global share and a strong U.S. footprint (Statista).
In FY2024, Imaging delivered roughly $10.6B in revenue, while Pharmaceutical Diagnostics contributed to just about $2.3B with ~18% YoY growth driven by procedure volumes and agent demand (FierceBiotech, BioSpace).
For comparison, here’s the market share of the top three diagnostic imaging vendors:
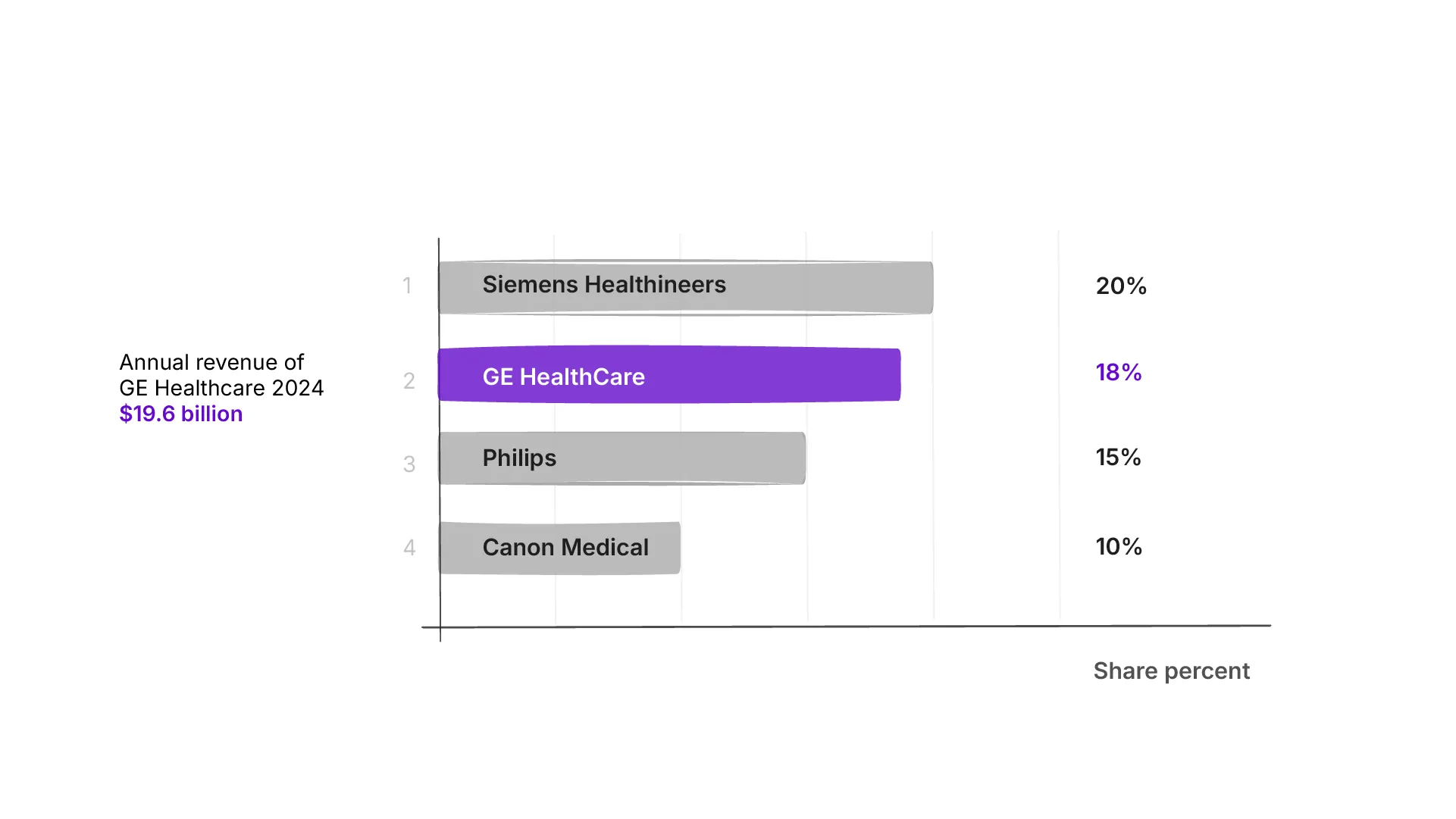
While Imaging remains the largest revenue driver, the integrated software and services layer, especially in oncology, compounds value across that hardware footprint. It is not the primary money maker, it is the multiplier.
OncoCare brings the oncology workflow into one workspace, unifying imaging, pathology, staging, and prior therapies.
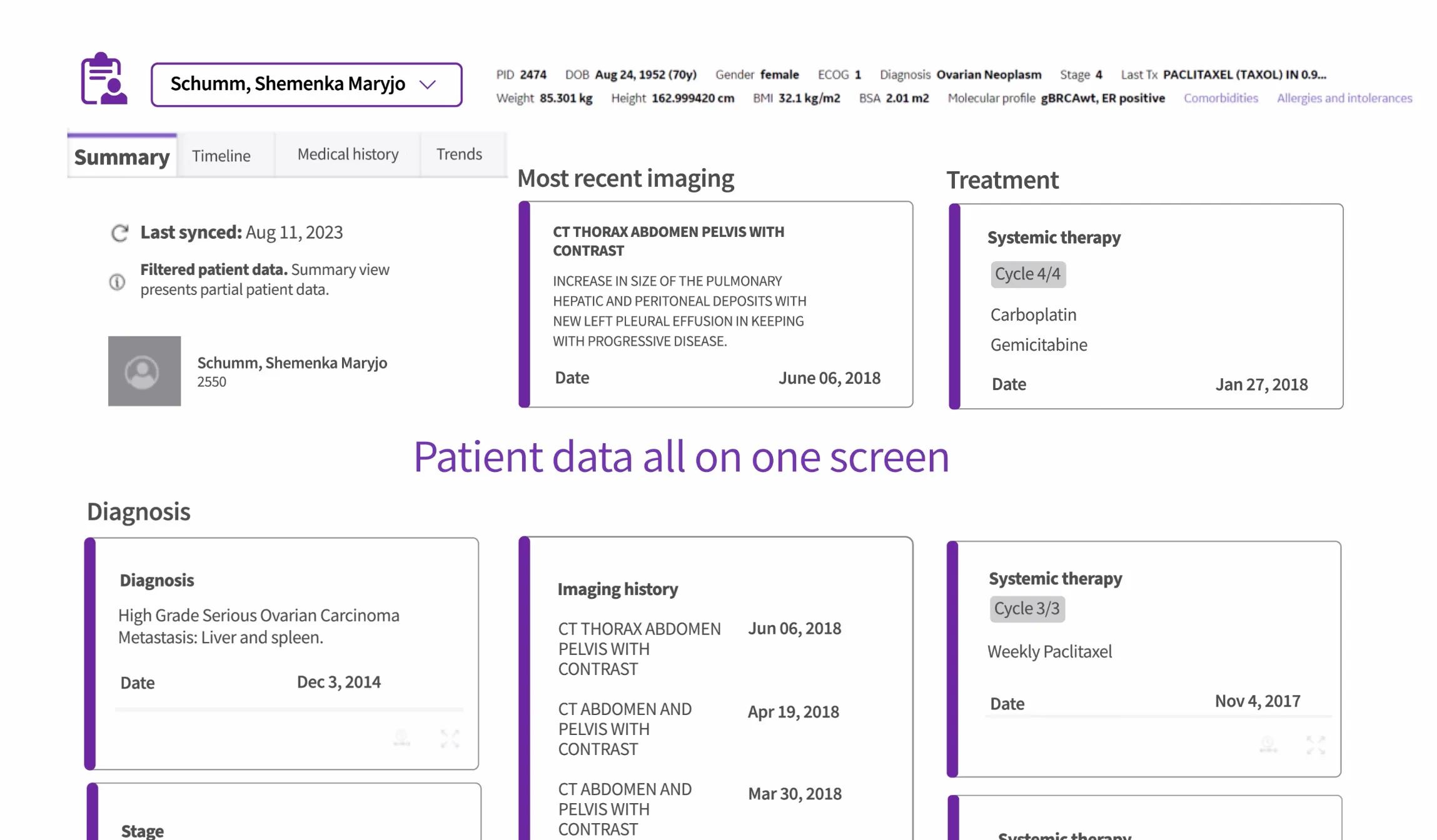
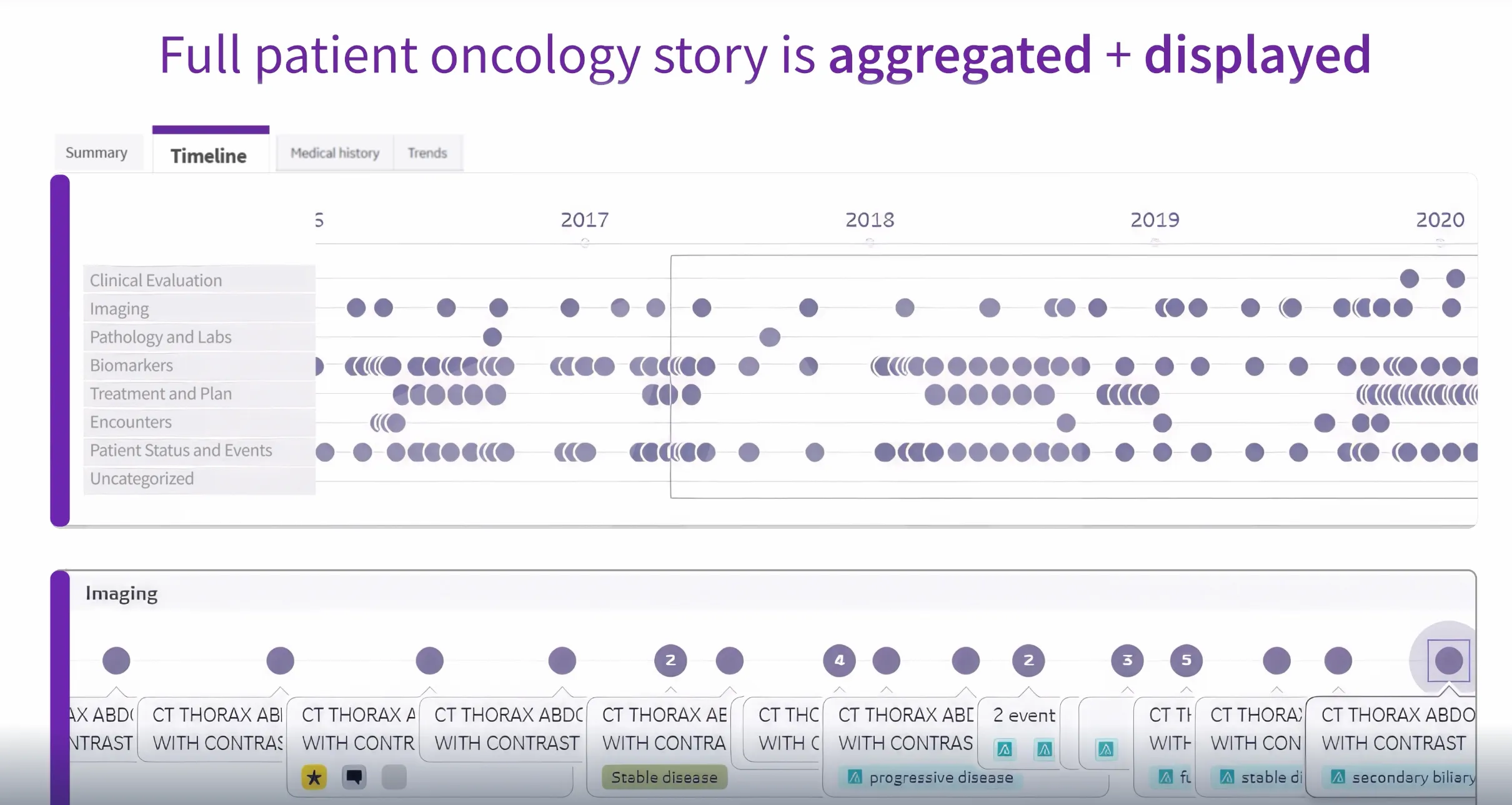
This alone adds immediate value to oncologists. Instead of opening multiple windows to reconstruct a patient’s history, they can see the longitudinal story in one place.
This type of application deployment is called a EHR Side-car application. The practitioner still continues to use the EHR, but the application sits on the side of the EHR, and the practitioner switches between the EHR and the application to make key decisions.
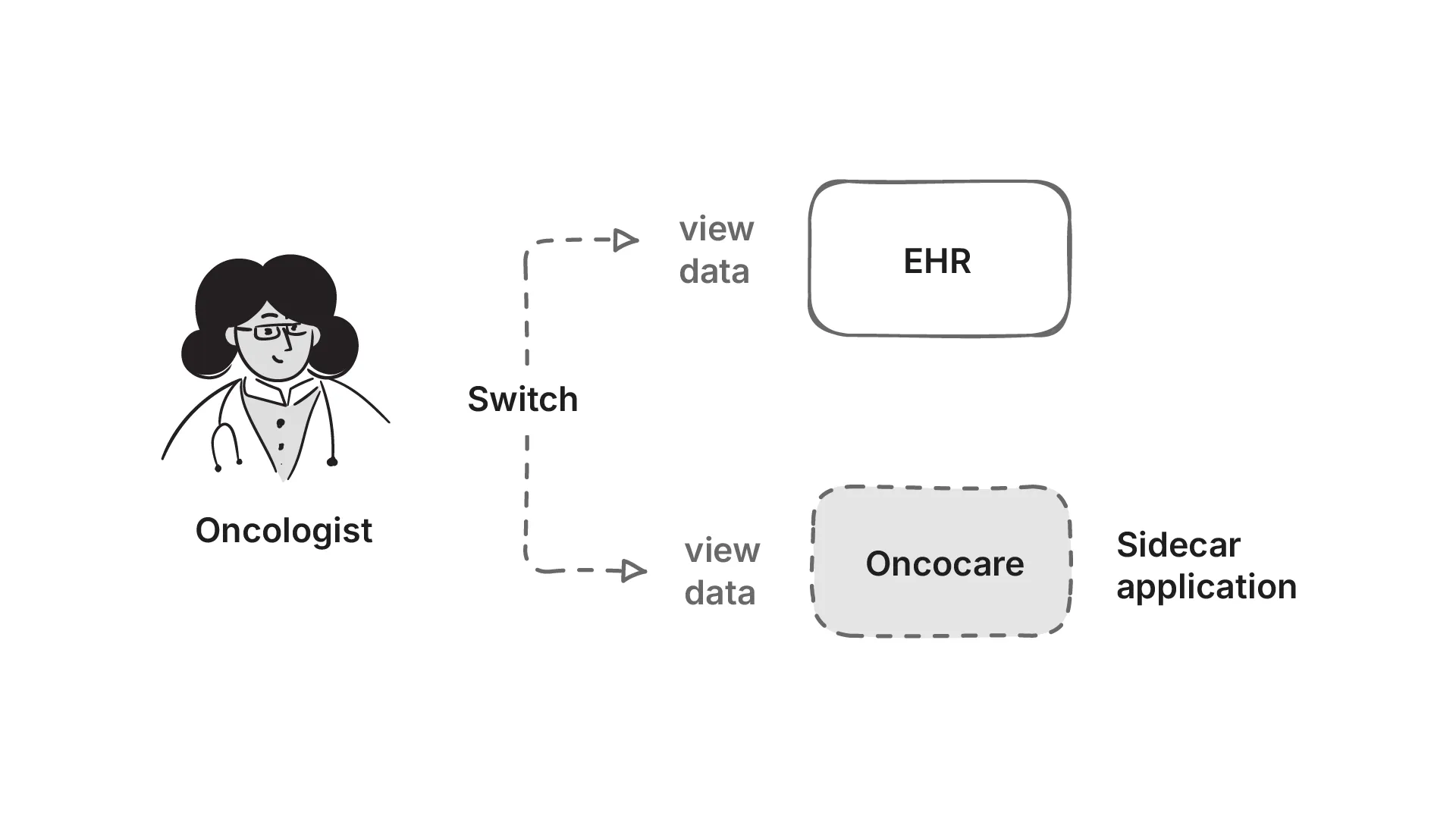
A sidecar is a separate system that runs alongside the main EHR. It doesn’t replace the EHR, but it adds capabilities that the EHR can’t provide on its own. Read more about it here.
Problem Statement
As an EHR sidecar application, OncoCare must stay in lock‑step with the EHR. This means syncing with the EHR’s data in real time. The ideal is SMART on FHIR EHR Launch with practitioner context, and pulling fresh data on demand on every launch, but many enterprise deployments either don’t expose it or restrict scopes (HL7 SMART App Launch). There is also an intense amount of data processing that needs to happen before the practitioner looks at OncoCare.
We therefore have to ingest from what is available: HL7 v2 ADT/ORM/ORU feeds (HL7 v2), C‑CDA documents (C‑CDA), DICOM metadata for imaging (DICOM), and even OMOP extracts in research settings (OMOP). There was also FHIR Bulk APIs available in some EHRs. Each source lands with different timing and semantics, we’ll need to normalize identifiers and code systems before anything touches decision logic.
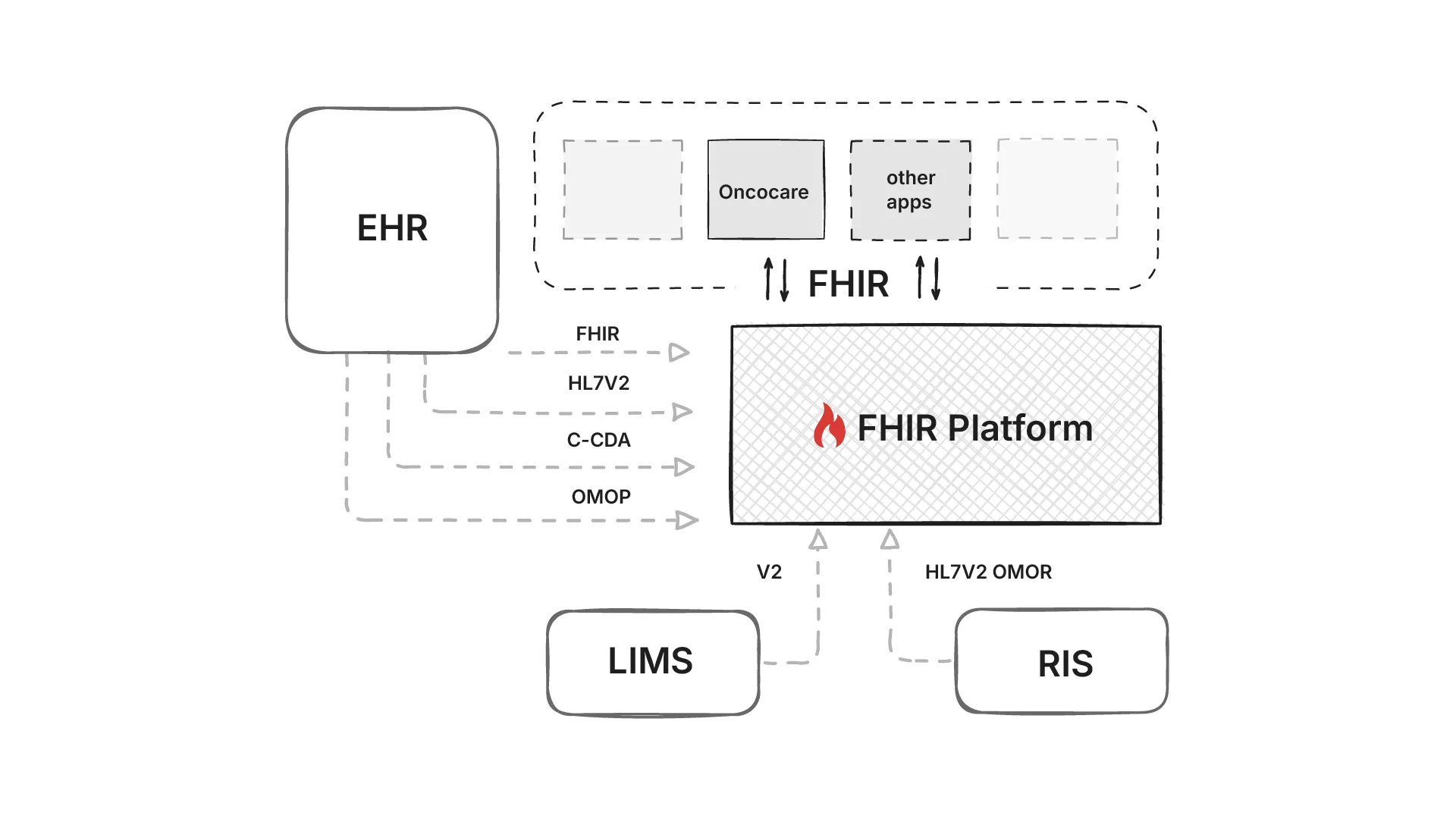
Because this standardization benefits every downstream app, not just OncoCare, it is placed on a common data platform. GE HealthCare’s Edison platform hosts clinical and AI applications. It exposes fully functional FHIR APIs - the hard part is transforming all those inputs into semantically consistent FHIR resources that the whole ecosystem can understand.
Problem statement 1: Get data from different formats into the Edison platform, as harmonized FHIR resource, fast
Next comes the hard part: converting the guidance by the National Comprehensive Cancer Network published as PDFs into computable rules that can be used by OncoCare for Clinical Decision Support (CDS). These rules also need to run on the FHIR data platform using standard FHIR APIs.
Although there has been a push for computable guidelines worldwide (like HEDIS pushing CQL, HQMF, and openEHR’s GDL2) the NCCN guidance is authored as human‑readable PDFs, with no plans to publish computable content.
We needed a faithful translation that preserves intent and can run on the FHIR resources we’ve already built.
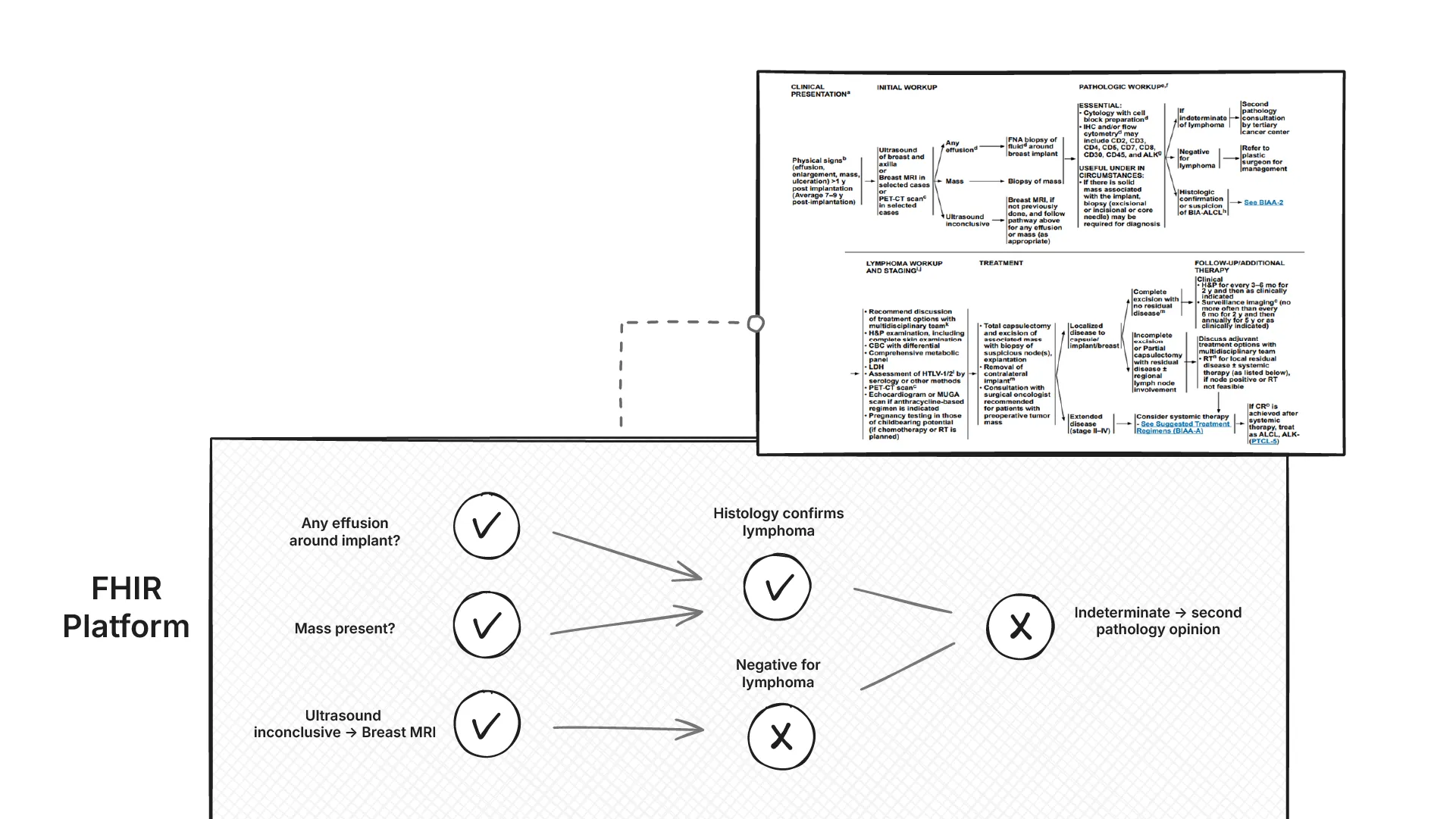
Explainability of these rules is non‑negotiable. For each recommendation, we not only need to return what the current recommendation is, but also the exact data used to trigger the decision (with codes and provenance), and why alternatives did not apply. Outputs are to be rendered as clinician‑friendly workflow graphs.
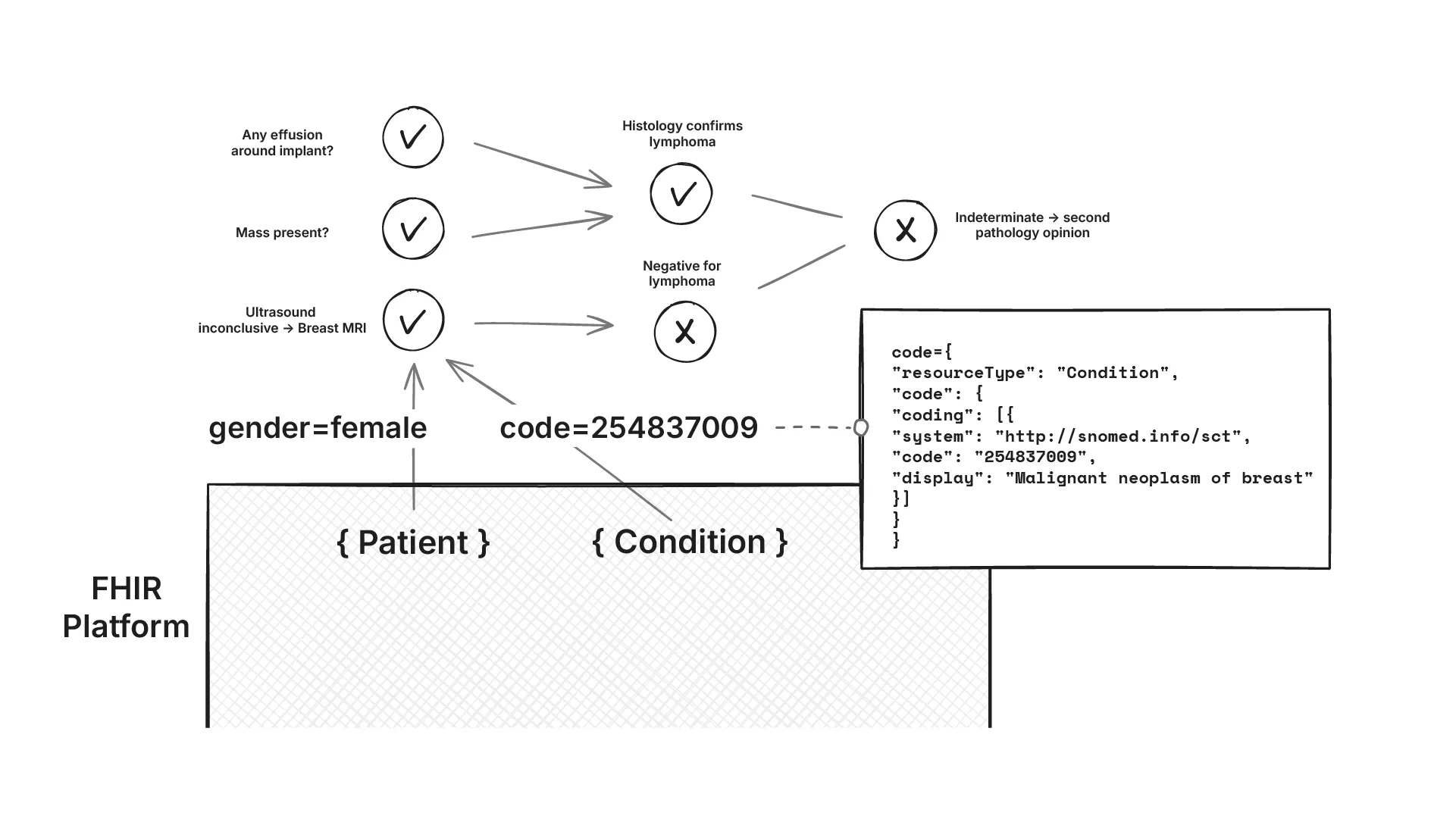
Problem statement 2: Convert NCCN guidance into computable CDS rules that run on the FHIR data platform, and are explainable to the clinician.
Harmonization of data into FHIR resources
The hardest part of any health data platform is getting data from multiple sources to speak the same language. In our case, we needed to transform data from FHIR Bulk APIs, OMOP research datasets, C-CDA documents, and HL7 v2 feeds into a unified FHIR representation that every GE HealthCare Edison application could understand.
The Challenge: Multiple Formats, Same Concepts
Consider how tumor size gets represented across different systems:
- OMOP: Stored in the
measurementtable with a concept_id pointing to LOINC 33728-7 (“Size.maximum dimension in Tumor”) - Epic FHIR Bulk: Comes as an
Observationresource with LOINC 21889-1 (“Size Tumor”) - Cerner FHIR Bulk: Uses
Condition.stage.assessmentwith a proprietary code system - C-CDA: Buried in unstructured text within a
<text>element of an observation
Each source was technically correct, but they couldn’t talk to each other. The FHIR platform needed one consistent way to represent tumor size so that downstream applications like OncoCare could reliably find and use this data.
Choosing the Right FHIR Profile
We initially considered using mCODE (Minimal Common Oncology Data Elements), which defines cancer-specific FHIR profiles. mCODE has a dedicated PrimaryTumorSize profile that would have been perfect for our tumor size example.
However, we hit a practical problem. Most applications already built on the Edison platform expected US Core FHIR profiles. These applications were looking for basic Observation resources, not specialized mCODE profiles. Switching to mCODE would have broken existing integrations.
We made a pragmatic choice: normalize everything to US Core profiles first, then use FHIR extensions to capture oncology-specific nuances that US Core couldn’t handle.
Our Harmonization Approach
Here’s an example of how we transformed that tumor size data:
From OMOP measurement table:
SELECT
measurement_concept_id, -- LOINC 33728-7
value_as_number, -- 7.0
unit_concept_id -- UCUM cm
FROM measurement
WHERE person_id = 12345To US Core Observation:
{
"resourceType": "Observation",
"code": {
"coding": [{
"system": "http://loinc.org",
"code": "33728-7",
"display": "Size.maximum dimension in Tumor"
}]
},
"valueQuantity": {
"value": 7.0,
"unit": "cm",
"system": "http://unitsofmeasure.org"
},
"subject": {
"reference": "Patient/12345"
}
}The transformation looks simple, but the devil was in the details. We had to maintain mappings between different OMOP concept IDs, various LOINC codes that represent the same concept in FHIR. Some EHRs used different LOINC codes for the same concept, and others used internal codes that we had to reverse-engineer.
Different Representations of the Same Concept Across EHRs
Even when two EHRs both claimed to support FHIR, their data looked different. Epic might represent a breast cancer diagnosis as:
{
"resourceType": "Condition",
"code": {
"coding": [{
"system": "http://snomed.info/sct",
"code": "254837009",
"display": "Malignant neoplasm of breast"
}]
}
}While Cerner used something like:
{
"resourceType": "Condition",
"code": {
"coding": [{
"system": "http://hl7.org/fhir/sid/icd-10-cm",
"code": "C50.9",
"display": "Malignant neoplasm of unspecified site of unspecified female breast"
}]
}
}Of course, the reality is more complex, and there are multiple codes for the same concept.
All representations were technically correct, but applications expecting SNOMED codes couldn’t understand ICD-10, and vice versa. We will revisit this later when building the decision support rules. For the time being, we directly integrated the data into the FHIR data platform as is.
Extensions for Oncology-Specific Data
Some oncology concepts simply don’t fit into standard US Core profiles. Take tumor staging - it involves multiple interrelated components (T, N, M stages) that don’t map cleanly to a single Observation.
We created extensions to capture this complexity:
{
"resourceType": "Observation",
"code": {
"coding": [{
"system": "http://loinc.org",
"code": "21908-9",
"display": "Stage group.clinical Cancer"
}]
},
"valueCodeableConcept": {
"coding": [{
"system": "http://cancerstaging.org",
"code": "3C",
"display": "Stage IIIC"
}]
},
"extension": [{
"url": "http://ge.com/fhir/extensions/tnm-staging",
"extension": [
{
"url": "t-stage",
"valueCodeableConcept": {
"coding": [{
"system": "http://cancerstaging.org",
"code": "T2",
"display": "T2"
}]
}
},
{
"url": "n-stage",
"valueCodeableConcept": {
"coding": [{
"system": "http://cancerstaging.org",
"code": "N1",
"display": "N1"
}]
}
}
]
}]
}This approach let us stay compatible with existing applications already using the FHIR platform while still providing the rich oncology data that OncoCare needed for decision support.
Decision support on FHIR APIs
Now came the harder problem: converting NCCN guidelines into computable rules that could run on our FHIR data platform. The challenge wasn’t just translation - it was dealing with the messy reality of how clinical data gets represented.
The Data Ambiguity Problem
Clinical concepts don’t map cleanly to FHIR resources. Take tumor size - it might appear as:
- A
Condition.stage.assessmentreference - An
Observationwith a LOINC code - Buried in unstructured text within a
DocumentReference
The same applied to medications. A patient’s current chemotherapy regimen could be scattered across MedicationRequest (what was ordered), MedicationStatement (what the patient reported), and MedicationAdministration (what was actually given). Our rules needed to check all three to get the complete picture.
Building the Rules Engine
We initially tried Open Policy Agent with Rego for rule evaluation. OPA provided good explainability and HTTP caching prevented redundant rule executions. But performance became an issue as rule complexity grew.
We ended up building our own rules engine with aggressive optimization. Instead of evaluating rules on-demand, we pre-computed decisions and only triggered re-evaluation when the underlying FHIR data changed. This reduced response times from seconds to milliseconds.
Terminology as the Foundation
The most challenging part was creating and maintaining terminology value sets. A single clinical concept like “breast cancer” might be represented by dozens of SNOMED CT, ICD-10, or proprietary codes across different EHRs.
We built value sets that captured these variations:
Breast Cancer Value Set:
- SNOMED: 254837009 (Malignant neoplasm of breast)
- SNOMED: 408643008 (Infiltrating duct carcinoma)
- ICD-10: C50.9 (Malignant neoplasm of breast, unspecified)
- Epic: 123456 (Breast CA)
- Cerner: BC001 (Breast carcinoma)Rules could then query against the value set rather than individual codes, making them resilient to EHR variations.
Explainable Decision Workflows
For each recommendation, we generated visual workflow graphs showing exactly which data triggered which decisions. Activated nodes highlighted the path through the NCCN logic, making the reasoning transparent to clinicians.
Each rule was tagged with its source NCCN guideline page number. When a rule fired, clinicians could see both the recommendation and the specific guideline section that justified it. This built trust and enabled quick verification against the original guidelines.

Since our engine used computable content, hospitals could also add their own institution-specific rules that weren’t yet published in NCCN guidelines.
Results
We successfully delivered both problem statements: a unified FHIR data platform that harmonized multiple data sources, and explainable clinical decision support that converted NCCN guidelines into computable rules.
The system processed data from Epic, Cerner, OMOP research datasets, and legacy HL7 v2 feeds into consistent US Core FHIR resources. Our rules engine evaluated NCCN decision points with sub-second response times, providing visual workflow explanations for each recommendation.
The pilot deployment demonstrated the system in one of U.S. healthcare’s largest conferences. Oncologists could see patient timelines, staging information, and treatment recommendations with full traceability back to NCCN guidelines, and many were blown away by the functionality.
Medblocks was a surprising find. We were looking for someone who had plug-and-play FHIR expertise. We needed to adjust from a lab environment to the real world. I was happy we could leverage your speed, and we didn’t need to tell you everything - you guys brought your own perspective and solved problems in your own way, which was very well accepted. It was a good experience overall, which is why we are looking at how to continue doing more work with Medblocks
- Ravi Bharadwaj, Senior Principal Software Engineer at GE HealthCare
Conclusion
This project reinforced several key lessons about health data platforms and clinical decision support. Some of these are:
- Data harmonization is always the hardest part. We spent 70% of our effort getting data from different sources to speak the same language. Every EHR implements FHIR differently, even when they claim compliance. The semantic gap between clinical concepts and transport formats requires constant mapping and translation work.
- FHIR as a semantic layer has limitations. While FHIR provides a common structure, it doesn’t solve the fundamental problem of different code systems representing the same clinical concepts. Terminology management becomes the critical bottleneck for any serious clinical application.
- Explainability builds trust. Oncologists didn’t just want recommendations - they wanted to see the reasoning. Visual workflow graphs and guideline references were essential for clinical acceptance. Black-box AI recommendations, no matter how accurate, wouldn’t have worked in this context.
- Performance matters for real-time decisions. Pre-computing decisions and triggering updates only when data changes was crucial for sub-second response times. Traditional rule engines that evaluate everything on-demand don’t scale for complex clinical decision support.
The combination of unified data platforms and explainable decision support creates a powerful foundation for clinical applications. But the devil remains in the details of data harmonization and terminology management.
Have a Health IT project in mind? Let’s talk about how we can make it happen.

