Interoperability doesn’t fail for lack of principles; it fails when there’s a lack of packages.
We discuss this and more in the latest episode of the Digital Health Hackers podcast, Dr. Sidharth Ramesh interviews two very interesting voices,
- Daniel Kapitan, a data scientist based in Rotterdam with over 25 years of experience. Daniel’s work in the past decade has focused on health data interoperability and data architecture.
- Joost Holslag, a medical doctor turned Health Informatics practitioner who works as an independent medical informatics consultant. Joost specializes in data modelling and openEHR.
A recent paper titled ‘Converge or Collide? Making Sense of a Plethora of Open Standards in Health Care’ framed this discussion. The paper is authored by leaders from leading standards communities including, Guy Tsafnat from OMOP, Rachel Dunscombe from openEHR,and Grahame Grieve from FHIR. The paper proposed that each of the standards, OMOP, openEHR and FHIR were needed, and catered to separate domains.
You can find a video discussion here,
This paper proposed that different data standards represented different points of view, and responded to different needs. It also stated that no single standard would necessarily be able to meet all requirements of healthcare systems. It concludes that the three open standards under discussion, OMOP, openEHR and FHIR were particularly suited to different healthcare domains, as represented in the below diagram.
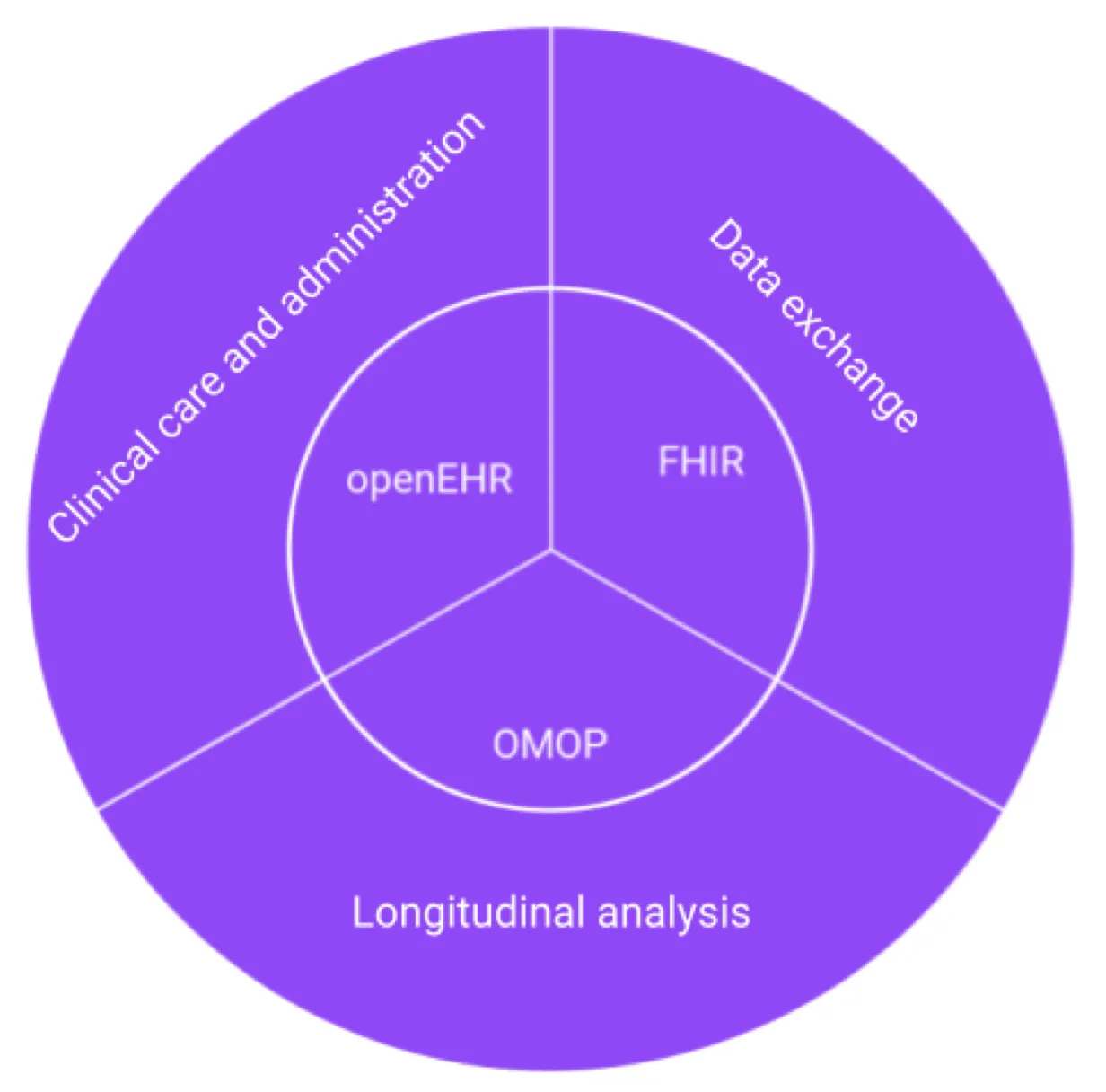
It recommends,
- openEHR for clinical care and administration as it provides a common data structure for health IT systems, including EMR and Patient Administration systems
- FHIR for data exchange, as at the center of the FHIR spec is a RESTful APO that provides real world healthcare services
- OMOP for longitudinal analysis, as it uses a patient-centric relational model making it suitable for complex queries across many patient records
While a guide for the application for each of these standards is a good place to start, Daniel argues that the paper wasn’t specific enough to guide system design in practice. In his response titled, ‘Data Interoperability in Context: The Importance of Open-Source Implementations When Choosing Open Standards’ it is argued that ‘open standards are a necessary but not sufficient condition to achieve health data interoperability’. He draws on his recent work in Kenya and the Netherlands to describe how FHIR is often used in ways beyond the ‘data exchange’ role. So how do these ideas play out once you leave the page and enter the field?
From theory to practice
Daniel refers to two major efforts he’s been involved in over the last 3 years that has shaped his view.
In the Netherlands, Daniel worked on the The PLUGIN Healthcare Project. This is a nation-wide effort to support federated analytics and federated learning. The aim is to let organizations analyze health data without having to centralize it into one large repository. Here, FHIR was adopted both for exchange, and as a way to model and move data inside subsystems that need to operate across institutional boundaries.
In Kenya and Tanzania, partnering with the PharmAccess, Daniel’s team was involved in rolling out Health Information Exchanges (HIEs) based on the OpenHIE architecture. Facilities there often lacked even desktops, not to mention servers, and the challenge was to build the exchange step by step rather than as a single monolith.
Daniel found local startups in Nairobi already building FHIR-based. OpenSRP by Ona is explicitly FHIR-native and designed for frontline workers, with HAPI FHIR commonly used as the server. That local ecosystem and the availability of Google’s Open Health Stack (Android FHIR SDK, FHIR Analytics, etc.), made it easier to adopt FHIR end-to-end.
As a result, FHIR was used across three layers,
- As the system of record for point of care apps
- As the exchange format for interoperability between services
- As a warehousing substrate where bulk FHIR exports are fed into open-source data pipelines for analysis
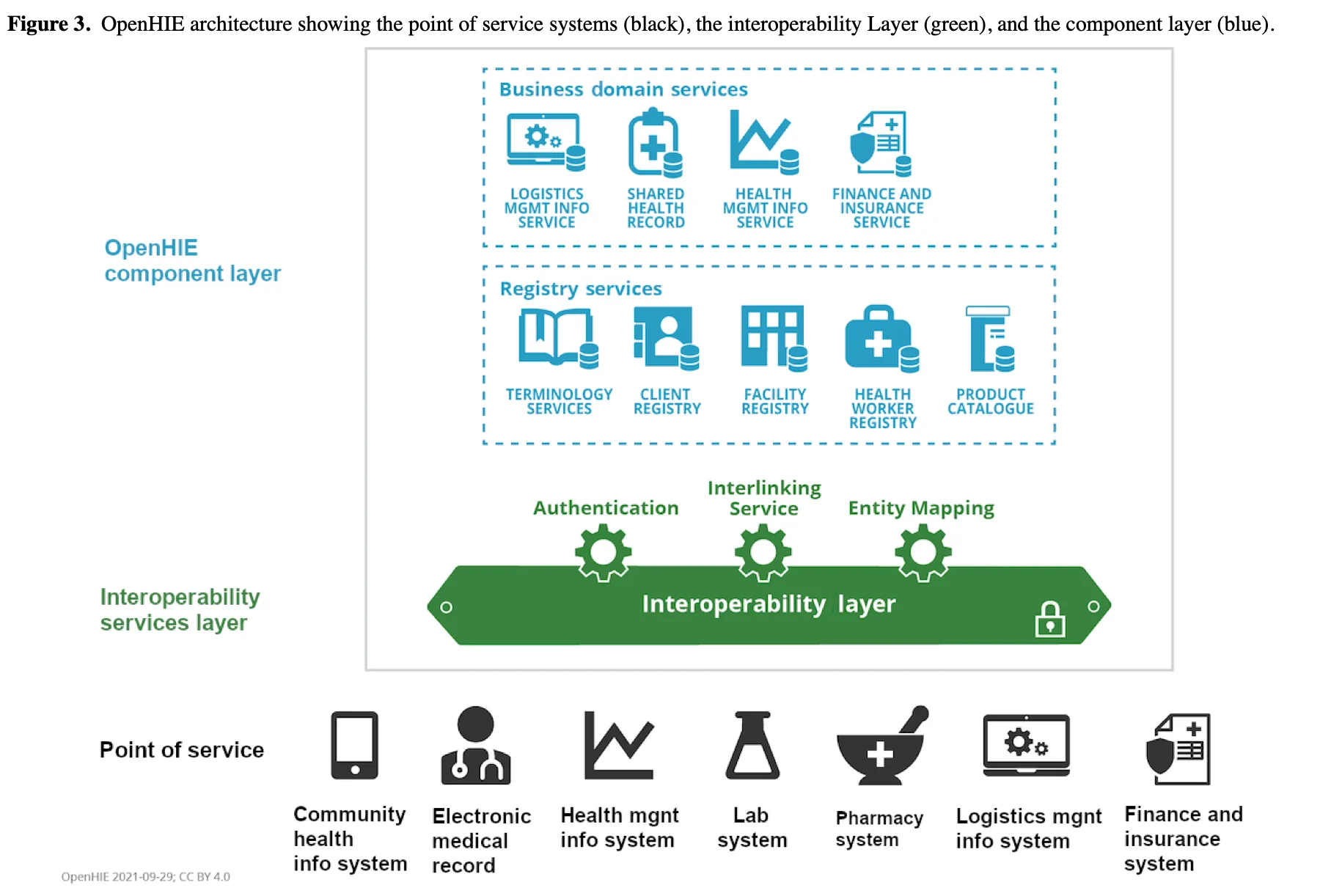
The takeaway here is that, once a standard is used one layer, (e.g. messaging), it often “leaks” into other layers(e.g. storage and analytics) beyond its intended use. This form of ‘standard leakage’ can happen because of developer familiarity, supportive ecosystems and in resource-constrained settings, it is easier to extend one standard than to train teams on many.
The accumulation of semantic debt
At this juncture, Joost offered a note of caution about what happens when FHIR is used across layers. FHIR is easy to adopt, but its ease of adoption often comes at the cost of consistency.
A shared health record on FHIR can hold patients, encounters, observations and conditions, and each of these concepts can be modelled in different ways. FHIR’s base specification doesn’t define concepts such as ‘blood pressure’ (unlike in openEHR). This means that the same concept can be modelled differently depending on the Implementation Guide it follows.
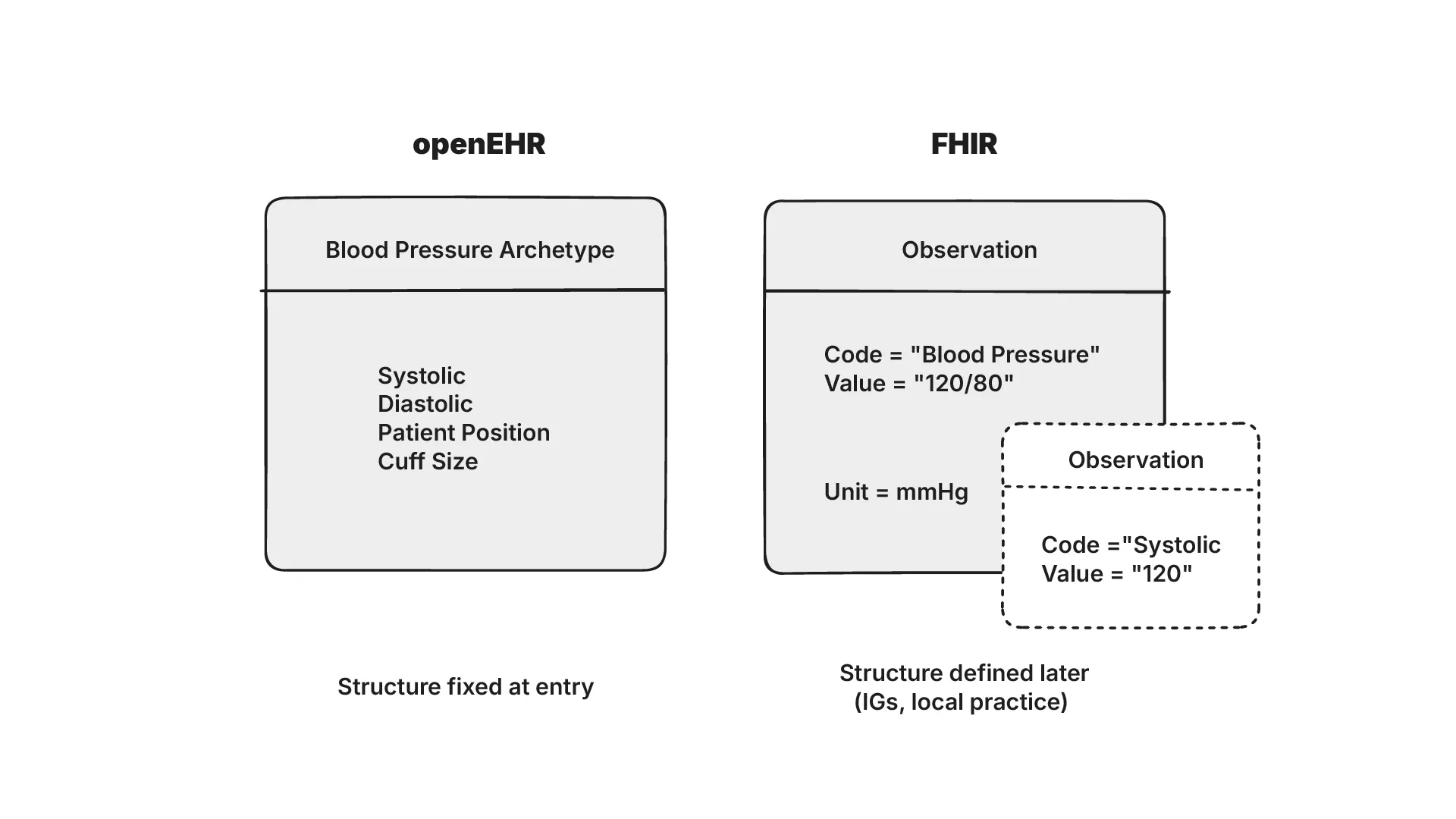
Joost describes this risk as ‘semantic debt’ where the low quality of initial data capture can lead to interoperability challenges as systems grow in complexity. Data that looks syntactically valid in FHIR may not be semantically comparable across systems. E.g. it may not be possible to “view all patients with fever” if “fever” isn’t defined consistently.
In the case of Daniel’s projects in Kenya, they turned to pragmatic workarounds to address this. For a maternal-care use case, simply by ensuring 4 clinic visits they could reduce mortality risks for mothers. Additionally they tracked only a few codeable concepts, including comorbidities like malaria and HIV. The approach they took involved writing transformations in SQL which was feasible since it only involved about ~100 ICD-10 codes.
Joost also uses the example of the lessons learned from the Dutch implementation, where billions were spent on integration with little value. This was caused by diverging Implementation Guides with different constraints. The experience has led Dutch stakeholders to shift towards adopting CKM archetypes as a stable semantic source of truth.
This conversation also highlighted a philosophical difference between FHIR and openEHR.
A philosophical difference: Early binding vs late binding
One way to understand the split between FHIR and openEHR is through the lens of when meaning is bound to data.
FHIR’s philosophy leans towards late binding. It assumes incoming data will be incomplete or inconsistent across systems. Rather than forcing strict alignment upfront, it wraps data in a resource and lets you exchange it anyway. Real binding happens later through implementation guides, mappings or downstream analytics. This lowers the barrier for adoption.
openEHR by contrast is built around early binding. It treats messy source data as the core problem, and insists on structure at the point of entry. Clinical concepts are formalized through archetypes and templates before capture, embedding meaning from the start. This does result in slower adoption, but ensures data is semantically stable and less prone to fragmentation.
Daniel brings up an interesting perspective here, his focus is usually on secondary use of data, that is, analyzing it after it has been recorded. Here, FHIR’s flexibility is an advantage. Joost’s perspective comes from his work in primary data entry and modelling where openEHR’s discipline is necessary.
Neither approach is right or wrong. The important question is to address where you want to confront complexity in your data lifecycle.
Transactional vs Analytical workloads
The confusion between shared health records and data warehouses (or transactional and analytical databases) is one that often appears in real implementations. On paper, this is how they are defined,
- A shared health record is a transactional system. It keeps track of individual patient data in real time and supports day-to-day clinical workflows.
- A data warehouse is an analytical system. It aggregates data across many patients and encounters to support dashboards, monitoring and population-level questions.
In practice, these roles are blurred. Questions asked by clinicians regularly require analytical workflows. E.g. “show me all patients who have not returned for a visit” requires scanning large portions of data that transactional systems aren’t designed to do efficiently.
Daniel explained that this was often misunderstood during his work in HIEs. The HAPI FHIR server used as a shared health record was sometimes assumed to be the data warehouse. His position on it was clear. For analytics, use bulk exports and move the data into systems built for purpose, such as SQL on FHIR and ClickHouse.
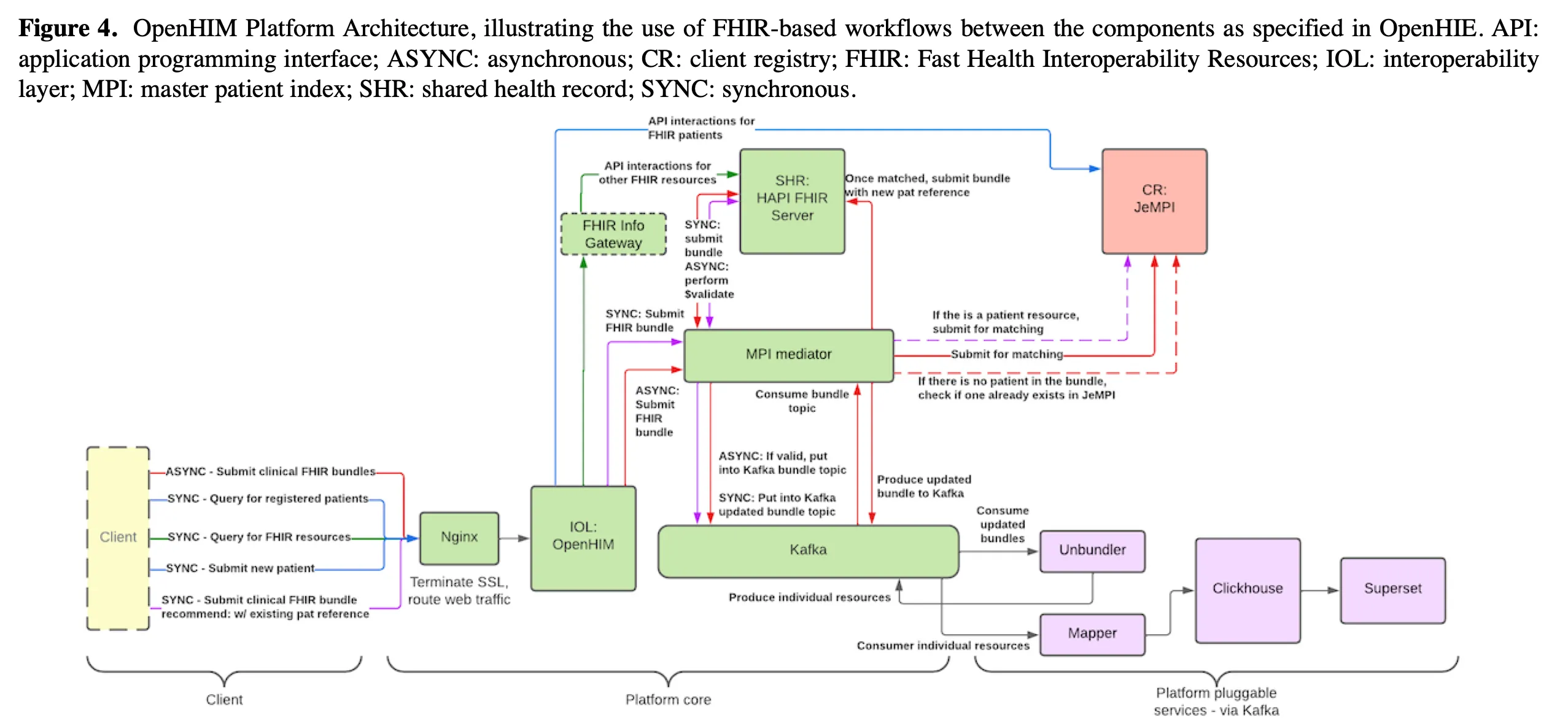
Separating transactional and analytical workloads isn’t optional according to Daniel. It reduced load on production systems and allowed analytical queries to be handled with the appropriate tools. In the long run, the shared health record could just as well be powered by openEHR (if the mappings were mature), but whatever the standard, the transactional and analytical layers needed to be distinct.
Performance and the database debate
Once analytical and transactional systems have been defined, the next question is about performance. Both FHIR and openEHR often store their data as JSONB in Postgres. While it is a flexible approach, it does have its issues. Queries that chain multiple parameters or filters can quickly become expensive.
This however, shouldn’t come as a surprise. Sidharth cited the classic work, ‘What Goes Around Comes Around’ by Michael Stonebraker to establish that relational databases have been the gold standard for decades when it came to concurrency control and predictable performance. A more recent paper by Stonebraker, ‘What Goes Around Comes Around… And Around…’ analyzes existing database trends including graph, vector and key-value stores and comes to the same conclusion.
We predict that what goes around with databases will continue to come around in upcoming decades. Another wave of developers will claim that SQL and the RM are insufficient for emerging application domains. […] We caution developers to learn from history. In other words, stand on the shoulders of those who came before and not on their toes. One of us will likely still be alive and out on bail in two decades, and thus fully expects to write a follow-up to this paper in 2044.
There is, however, room for experimentation. Daniel expresses the potential he sees in graph databases which can be both flexible and expressive. They also discuss event-sourced or time-series databases that naturally capture changes over time. Daniel stresses that new approaches don’t mean abandoning old lessons. “Thou shalt make composable components,” he says, indicating his view of the future.
The answer lies less in finding the “perfect database” and more in using the right tool for the right job, relational for transactions, columnar for analytics, and graph or temporal models where flexibility or history matter most. Daniel extends his point beyond databases to system architecture as a whole. Policymakers are often tempted to ask for one national platform that holds everything. In his view this creates fragility, makes it harder to secure, scale and more vulnerable to political bottlenecks.
On the benchmarking side, evidence is mixed. A Smile CDR benchmark demonstrated ingestion performance of over 11k FHIR resources per second on AWS infrastructure with optimized configurations supporting tens of thousands of concurrent requests. In a separate large scale deployment, Smile Digital Health reports handling ~255k interactions/second while onboarding FHIR resources for 2M patients in ~26 hours. At the hyperscale end, Google Cloud Healthcare API has been shown to sustain 50M synthetic patient records (~26B resources) with linear performance, achieving per resource response times under 200 ms.
Joost notes that independent, comparable benchmarks remain scarce. Without shared and transparent benchmarks, it’s hard to know whether FHIR or openEHR repositories can really scale to the demands placed on them.
Can AI solve the problem of mappings?
And no podcast feels complete without discussing AI. In this case the focus was on how large language models might help with the necessary work of mapping data between systems. This could be through the blackbox integration of LLM written code that has been tested on multiple inputs and outputs, and then reviewed by a human.
Several perspectives emerged. Using LLMs as coding assistants to draft mappings in JSON or YAML quicker, with a human reviewer in the loop was the approach suggested by Daniel. Joost was skeptical about using LLMs at all as they are probabilistic and generate likely answers not correct ones. In clinical contexts, that uncertainty carries risk.
Daniel used the example of aviation, autopiloting technology was accompanied by an entire safety culture to ensure human vigilance. Health IT on the other hand, doesn’t have the same safeguards.
At the root of this conversation lies the fact that sometimes there isn’t a single ‘right’ mapping at all. The mapping is heavily dependent on the question to be asked and context to be considered. This makes it harder to automate the task completely.
The role for AI looks modest, but useful. Use LLMs to speed up the mechanics of writing transformations, but leave the responsibility for correctness with clinicians and informaticians.
Conclusion
This conversation traced an interesting path, from fieldwork to philosophical differences and architectural decisions. Daniel’s experiences in the Netherlands and Kenya showed how the neat domain split between openEHR, FHIR and OMOP blurs in practice. In resource constrained settings, the easiest way forward is to use what you know, and what has an open-source ecosystem. We also discussed how standard leakage means once a format works in one layer, it inevitably seeps into others.
Two dominant philosophies emerged, FHIR’s pragmatism in accepting messy data to enable fast adoption, and openEHR’s rigor in enforcing structure at entry to ensure long term stability. It is important to acknowledge the limitations of each, and decide where to tackle this complexity. Database choices and performance requirements point to the same principle, that there is no single perfect solution. Composable components in composable architectures seem to be the scalable way to go.
Looking forward, both Daniel and Joost agree on convergence rather than fragmentation. They stress the importance of shared semantics, with the International Patient Summary (IPS) as a potential starting point.
In the end, the message is clear. Open standards are necessary, but insufficient on their own. True interoperability requires more than specs, it needs governance, open source implementations and a commitment for standards communities to work together. Aligning on shared models and backing them up with open tooling will go a long way towards achieving health data interoperability.
Edited (5 Sep 2025): Edited to reflect feedback shared by Daniel Kapitan in the comments below

